GOTO NEW VERSION!) Version is 3.6 [Fall 2013]
Updated prior system, including a gamma distribution prior. Addition of prior information into the histograms of the PDF outfile. MacOS 106, 10.7, and 10.8 version now have an experimental installer. I switched the copyright license to the MIT opensource license.
Important additional information to help run recent version of migrate:
-
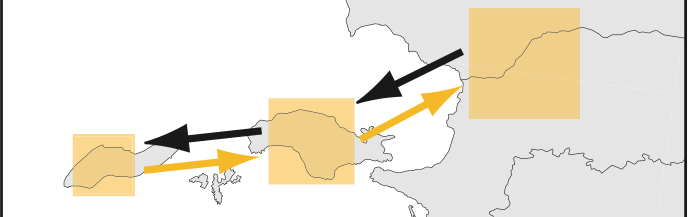
•Most recent paper on migrate technology: Beerli and Palczewski 2010: Unified Framework to Evaluate Panmixia and Migration Direction Among Multiple Sampling Locations. Genetics (2010) vol. 185 pp. 313--326 (LINK)
-
•Tutorial on how to compare population genetic models (LINK)
-
•Opinion about issues with divergence and accuracy of migration rates (LINK)
Known problems with current version:
-
• no known problem (yet)